-Search query
-Search result
Showing 1 - 50 of 764 items for (author: au & gg)

EMDB-43931: 
CryoEM structure of activated CRAF/MEK/14-3-3 complex with NST-628
Method: single particle / : Quade B, Cohen SE, Huang X

EMDB-43932: 
Activated CRAF/MEK heterotetramer from focused refinement of CRAF/MEK/14-3-3 complex
Method: single particle / : Quade B, Cohen SE, Huang X

PDB-9axa: 
CryoEM structure of activated CRAF/MEK/14-3-3 complex with NST-628
Method: single particle / : Quade B, Cohen SE, Huang X

PDB-9axc: 
Activated CRAF/MEK heterotetramer from focused refinement of CRAF/MEK/14-3-3 complex
Method: single particle / : Quade B, Cohen SE, Huang X

EMDB-41907: 
Computationally Designed, Expandable O4 Octahedral Handshake Nanocage
Method: single particle / : Weidle C, Borst A
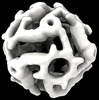
EMDB-42031: 
Computational Designed Nanocage O43_129_+8
Method: single particle / : Weidle C, Kibler RD

EMDB-43318: 
Twistless helix 12 repeat ring design R12B
Method: single particle / : Calise SJ, Kollman JM

EMDB-29974: 
Cryo-EM structure of synthetic tetrameric building block sC4
Method: single particle / : Redler RL, Huddy TF, Hsia Y, Baker D, Ekiert D, Bhabha G

EMDB-41364: 
CryoEM Structure of a Computationally Designed T3 Tetrahedral Nanocage
Method: single particle / : Weidle C, Borst AJ

EMDB-42906: 
Computational Designed Nanocage O43_129
Method: single particle / : Weidle C, Kibler RD

EMDB-42944: 
Computational Designed Nanocage O43_129_+4
Method: single particle / : Carr KD, Weidle C, Borst AJ

PDB-8gel: 
Cryo-EM structure of synthetic tetrameric building block sC4
Method: single particle / : Redler RL, Huddy TF, Hsia Y, Baker D, Ekiert D, Bhabha G

PDB-8tl7: 
CryoEM Structure of a Computationally Designed T3 Tetrahedral Nanocage
Method: single particle / : Weidle C, Borst AJ

PDB-8v3b: 
Computational Designed Nanocage O43_129_+4
Method: single particle / : Carr KD, Weidle C, Borst AJ

EMDB-17597: 
cryo-EM structure of Doa10 in MSP1E3D1
Method: single particle / : Botsch JJ, Braeuning B, Schulman BA

EMDB-17608: 
cryo-EM structure of Doa10 with RING domain in MSP1E3D1
Method: single particle / : Botsch JJ, Braeuning B, Schulman BA

EMDB-17609: 
Low resolution map of Doa10 in MSP1E3D1
Method: single particle / : Botsch JJ, Schulman BA

EMDB-17610: 
Doa10 in MSP2N2
Method: single particle / : Botsch JJ, Schulman BA

PDB-8pd0: 
cryo-EM structure of Doa10 in MSP1E3D1
Method: single particle / : Botsch JJ, Braeuning B, Schulman BA

PDB-8pda: 
cryo-EM structure of Doa10 with RING domain in MSP1E3D1
Method: single particle / : Botsch JJ, Braeuning B, Schulman BA

EMDB-16127: 
Yeast 80S, ES7s delta, eIF5A, Stm1 containing
Method: single particle / : Dimitrova-Paternoga L, Paternoga H, Wilson DN

PDB-8bn3: 
Yeast 80S, ES7s delta, eIF5A, Stm1 containing
Method: single particle / : Dimitrova-Paternoga L, Paternoga H, Wilson DN

EMDB-17736: 
ATTRV20I amyloid fibril from hereditary ATTR amloidosis
Method: helical / : Steinebrei M, Schmidt M, Faendrich M

EMDB-17737: 
ATTRG47E amyloid fibril from hereditary ATTR amloidosis
Method: helical / : Steinebrei M, Schmidt M, Faendrich M

EMDB-17738: 
ATTRV122I amyloid fibril from hereditary ATTR amloidosis
Method: helical / : Steinebrei M, Schmidt M, Faendrich M

PDB-8pke: 
ATTRV20I amyloid fibril from hereditary ATTR amloidosis
Method: helical / : Steinebrei M, Schmidt M, Faendrich M

PDB-8pkf: 
ATTRG47E amyloid fibril from hereditary ATTR amloidosis
Method: helical / : Steinebrei M, Schmidt M, Faendrich M

PDB-8pkg: 
ATTRV122I amyloid fibril from hereditary ATTR amloidosis
Method: helical / : Steinebrei M, Schmidt M, Faendrich M

EMDB-28850: 
SARS-CoV-2 Gamma 6P Mut7 S + COVA309-3 Fab
Method: single particle / : Torres JL, Ward AB

EMDB-28851: 
SARS-CoV-2 Gamma 6P Mut7 S + COVA309-10 Fab
Method: single particle / : Torres JL, Ward AB

EMDB-28852: 
SARS-CoV-2 Omicron 6P S + COVA309-35 Fab
Method: single particle / : Torres JL, Ward AB
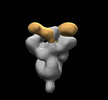
EMDB-28853: 
SARS-CoV-2 Gamma 6P Mut7 + S COVA309-38 Fab
Method: single particle / : Torres JL, Ward AB

EMDB-18191: 
X. laevis CMG dimer bound to dimeric DONSON - without ATPase
Method: single particle / : Cvetkovic MA, Costa A

EMDB-18192: 
X. laevis CMG dimer bound to dimeric DONSON - MCM ATPase
Method: single particle / : Cvetkovic MA, Costa A

EMDB-18195: 
Single CMG purified from replicating Xenopus egg extracts
Method: single particle / : Cvetkovic MA, Costa A

PDB-8q6o: 
X. laevis CMG dimer bound to dimeric DONSON - without ATPase
Method: single particle / : Butryn A, Cvetkovic MA, Costa A

PDB-8q6p: 
X. laevis CMG dimer bound to dimeric DONSON - MCM ATPase
Method: single particle / : Butryn A, Cvetkovic MA, Costa A

EMDB-29530: 
SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-29531: 
SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-40240: 
SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

PDB-8fxb: 
SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

PDB-8fxc: 
SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

PDB-8s9g: 
SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment
Method: single particle / : Park YJ, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler D

EMDB-28552: 
Human Membrane-bound O-acyltransferase 7
Method: single particle / : Wang K, Liao M, Farese RV, Walther TC

PDB-8erc: 
Human Membrane-bound O-acyltransferase 7
Method: single particle / : Wang K, Liao M, Farese RV, Walther TC

EMDB-29680: 
C6HR1_8r: Extendable repeat protein heptamer
Method: single particle / : Bethel NP, Borst AJ

EMDB-29847: 
C3HR3_8r: Extendable repeat protein trimer
Method: single particle / : Bethel NP, Borst AJ

EMDB-29849: 
C6HR1_4r: Extendable repeat protein hexamer
Method: single particle / : Bethel NP, Borst AJ

EMDB-29851: 
C4HR1_8r_shift5: Extendable repeat protein fiber
Method: helical / : Bethel NP, Borst AJ
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
